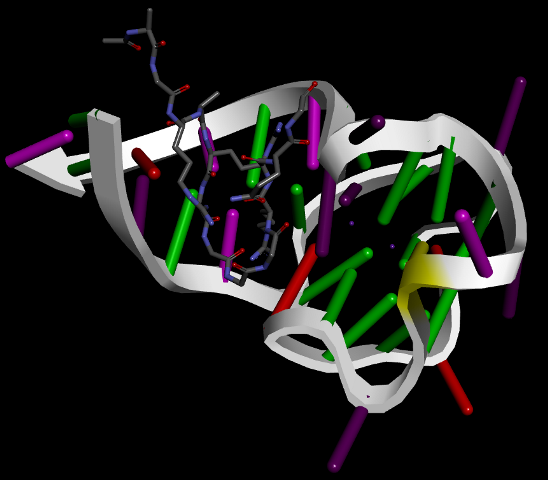
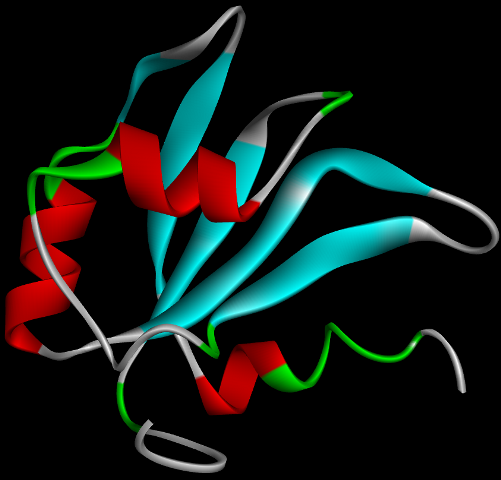
G4IPDB is an unique and interactive database of proteins that interact with G-quadruplex forming nucleic acid sequences.
This protein database contains the comprehensive information of nucleic acid targets, targets name, target sequences,
delta-Tm values, binding constant values, dissociation constant values, genes name, gene synonyms, FASTA sequences of proteins,
UniProt ID of proteins, nucleic acid interacting residues, PDB ID, interaction ID of proteins, the link for GeneBank database sequence graphical view, PMID, literature's authors name and the techniques used to study the interaction with their targets.
The link for protein's gene sequence graphical view is also available in our database.
The present database provides an efficient and robust tool to predict the putative G-quadruplex-forming sequences in the given sequences.
Database users have both options to enter the query sequence manually or browse and upload the query sequence directly from the local disk space in either FASTA or text file format.
This web-based tool predicts the putative G-quadruplex-forming sequences in both directions (sense and antisense strands) and provides a confidence score of cG/cC.
The uniqueness of the G4IPDB database is the assembled information on a single platform and user-friendly browsing.
G4IPDB database is freely available database which can be efficiently explored and searched using different proteins name and nucleic acid targets name.
To the best of our knowledge, G4IPDB is the first database that is specifically focused on any type of G-quadruplex DNA and RNA interacting protein and includes their interaction data as available in the literature.
We believe that the entries reported in this database would be useful for larger scientific community targeting the G-quadruplex nucleic acid and their binding protein as potential drug targets.their binding protein as potential drug targets.
Citation details : Mishra, S. K. et al. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins.
Sci. Rep. 6, 38144; doi: 10.1038/srep38144 (2016).